1. Introduction
Spinal muscular atrophy (SMA) is one of the most common lethal recessive genetic conditions, with an incidence of 1:8000 births in Europe dependent on ethnicity [
1,
2]. In the UK in 2019, there were 712,699 live births [
3] (UK Office for National Statistics), suggesting that approximately 71 babies were born that year with a type of SMA.
Until recently, the treatment for SMA was limited to supportive care, but in 2020, Nusinersen (an antisense oligonucleotide administered intrathecally) was approved for use in the UK by the National Institute for Health and Care Excellence (NICE). A second effective therapy (the adeno-associated viral vector onasemnogene abeparvovec) has been sanctioned for use in babies up to 12 months of age by NICE in 2021.
Since early diagnosis and intervention is crucial for therapy to be effective, there has been considerable global interest in newborn screening (NBS) for SMA. Many countries have now adopted NBS for SMA nationally or have embarked on pilot studies regionally [
4,
5,
6,
7,
8]. The methodology for SMA NBS has been fully validated elsewhere [
9] and is already utilised by many NBS labs today. However, no SMA NBS assay has been tested in the UK setting previously, and thus no NBS program for SMA in the UK exists despite a clinical need for early diagnosis.
In order to provide robust laboratory data within the UK setting, we completed a short study to test a widely used, commercially available assay on anonymised, presumed normal blood spots and known SMA-positive blood spots. We were thus able to assess the suitability and feasibility of running SMA newborn screening in a routine clinical laboratory focusing on assay performance at detecting SMA with minimal false positives, and hands-on time required by staff.
2. Materials and Methods
This study was approved by the London–West London and GTAC Research Ethics Committee (19/LO/1801). A total of 4810 anonymised leftover dried blood spots (DBSs) were retrieved from the newborn screening department at Great Ormond Street Hospital, London. In addition, 43 known anonymised SMA-positive dried blood spots from diagnosed SMA patients were provided by Biogen Idec Ltd. (Maidenhead, UK) and were dispersed randomly among the plate runs. The assay was carried out using the RUO NeoMDx DNA Extraction kit (PerkinElmer Wallac Oy, Finland) following the manufacturers’ instructions. In short, DNA was extracted from 3.2 mm punches taken from the dried blood spots and was eluted in 80 µL Elution Solution. Subsequently, 3 µL of this eluted DNA was used in a quantitative real-time PCR (rqPCR) with 12 µL of the manufacturer’s rqPCR mix that included primer and probes for SMN1 exon 7 and a control gene (RPP30). The rqPCR reactions were carried out on a Bio-Rad CFX PCR machine with the following amplification steps: 37 °C for 2 min and 94 °C for 5 min, followed by 50 cycles of 93 °C for 10 s, 60 °C for 30 s, and 69 °C for 40 s. Results were analysed using the Bio-Rad CFX Manager software (v3.1). Two sets of each kit control (C1, C2, and C3) were run with each plate. The C1 control contained no SMN1 gene, whilst the C2 and C3 controls contained normal wild-type SMN1 in an identical concentration.
3. Results
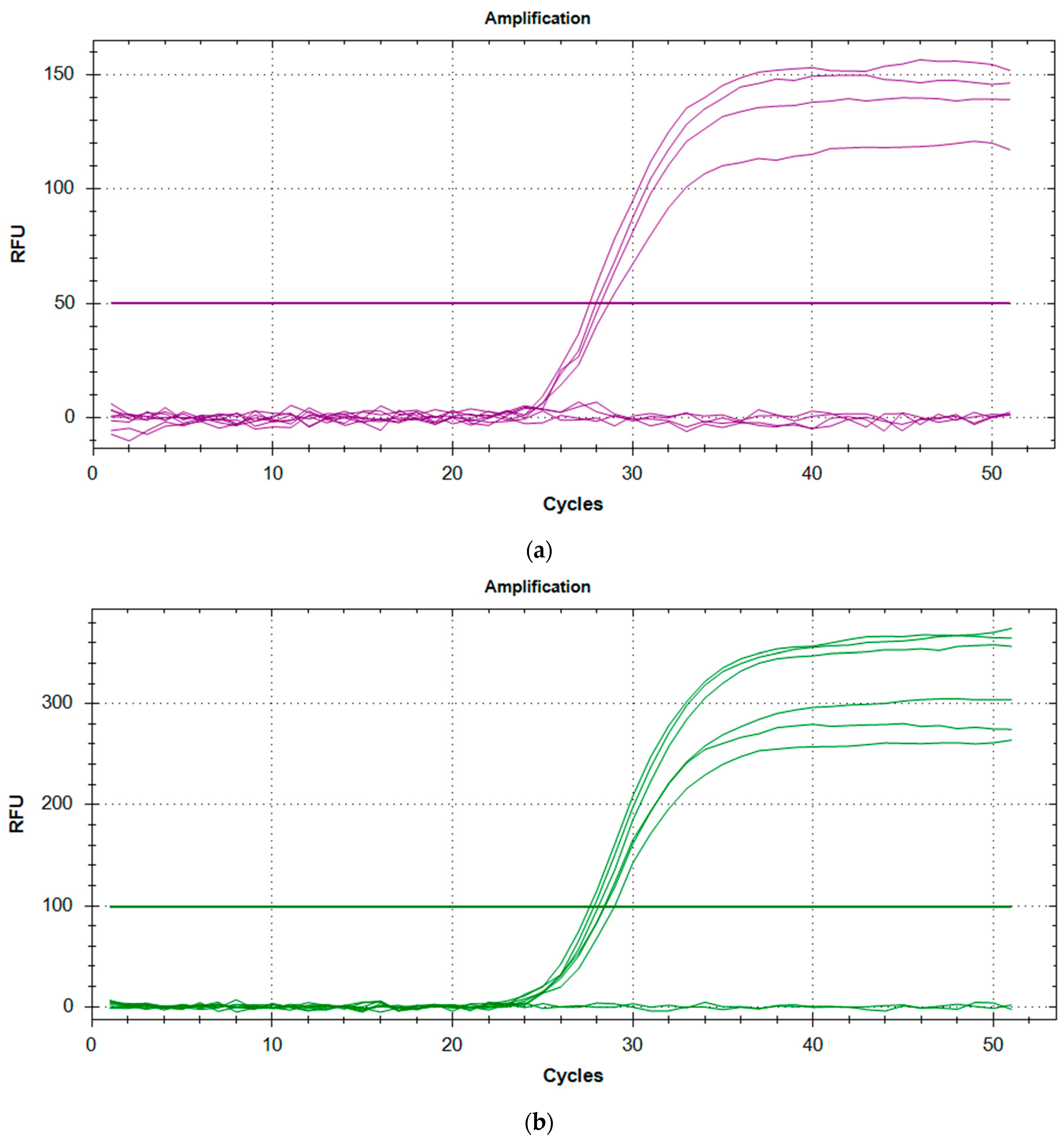
After initial staff training (data from training not included in these results or analysis), all the rqPCR runs were successful with the
RPP30 controls, the
SMN1 controls, and the blank samples demonstrating consistent and reproducible amplification (
Table 1,
Table 2 and
Figure 1). Controls and blanks were included in each run with typically one run carried out per day. This permitted reliable analysis of the anonymised, presumed normal DBS and the known SMA-positive DBS, as well as the kit controls provided in the NeoMDx PCR Reagent kit (PerkinElmer Wallac Oy). Since only one run was performed per day, the data shown in
Table 1 and
Table 2 illustrates interday precision for the kit controls run in duplicate. The results are within the 2 SD limits of acceptability for assay consistency and reproducibility.
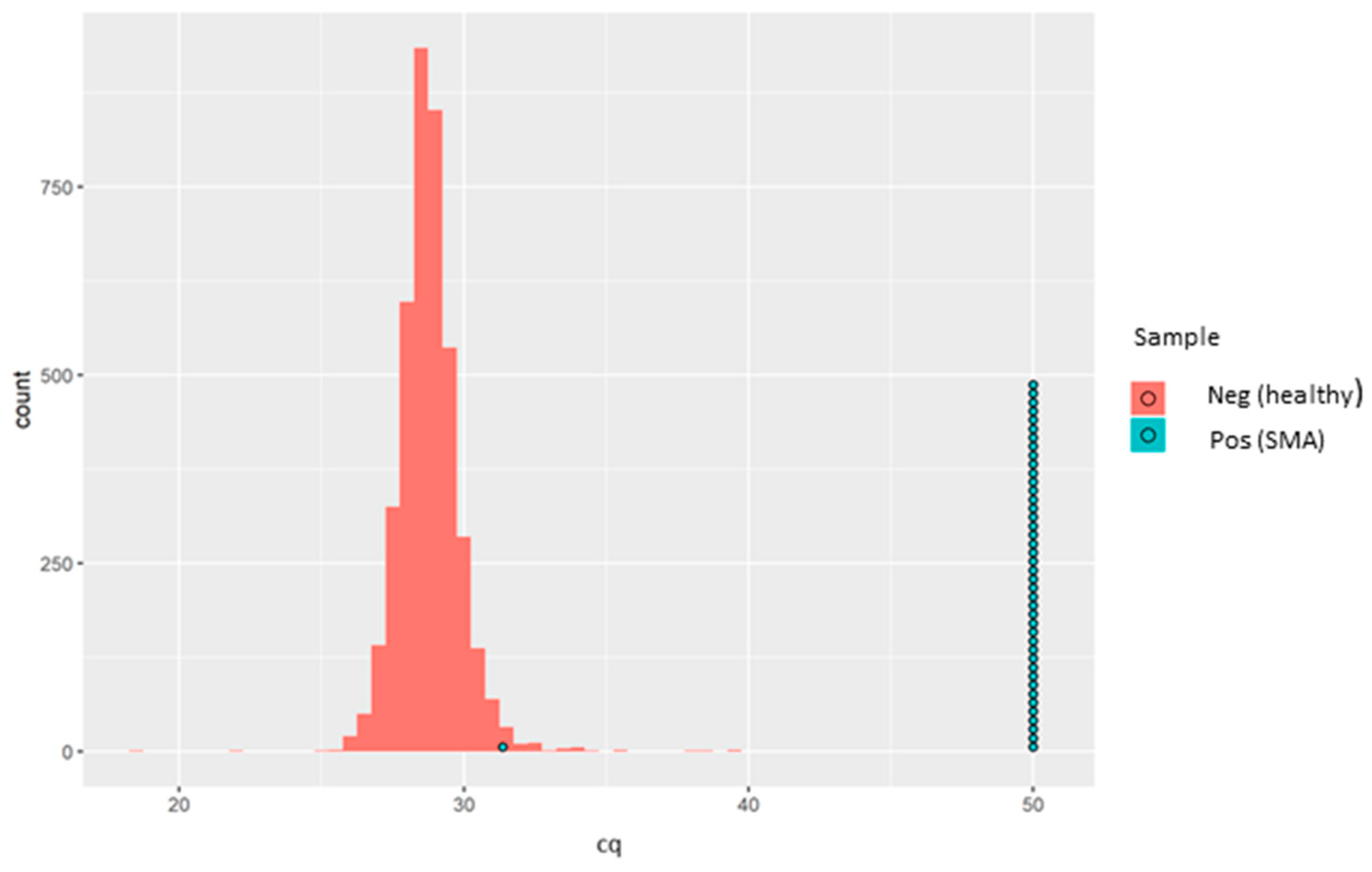
Since this is qualitative (i.e., absence or presence of
SMN1 exon 7), no quantitative controls were run, and thus no calibration curve was generated. All the 4810 anonymised, presumed normal DBS samples and all 43 of the known SMA-positive samples demonstrated positive amplification for the
RPP30 control gene. Meanwhile,
SMN1 amplification was detected in all the 4810 anonymised, presumed normal DBS samples. Of the 43 known SMA-positive DBS samples, 42 showed complete absence of
SMN1 amplification. Interestingly, a single SMA-positive sample showed clear positive amplification of the
SMN1 gene with an rqPCR cycle threshold (Cq) of 31.36 (
Figure 2).
It was initially presumed that this sample might have had an alternative variant in the
SMN1 gene not covered by the rqPCR assay. However, after contact with Biogen, who provided us in a blinded fashion the 43 samples with known genotypes, this “SMA positive” sample was actually derived from a carrier, and not from an affected individual, and therefore serendipitously acted as a good quality control for the assay since the aim was not to identify carriers. Thus, in our hands, this assay showed excellent analytical specificity for
SMN1 exon 7 deletion. The sensitivity of the
SMN1 amplification, based on Cq values distribution, is demonstrated in
Figure 2.
4. Discussion
Although this assay is already widely used in numerous NBS centres, SMA NBS has never been assessed in a UK laboratory setting before. In our hands, the assay has excellent specificity and successfully identified all SMA-positive samples, and did not identify a single SMA-positive case in the presumed normal, anonymised DBS. In addition, the sample initially erroneously described as SMA positive (subsequently found to be from a carrier) showed positive SMN1 amplification with the NeoMDx assay. This demonstrates that the assay only identifies individuals with homozygous exon 7 deletions. This is imperative for use as a screening tool for only SMA-affected individuals.
The test was easily adopted into practice in the UK clinical laboratory setting with minimal hands-on time for staff and a simple, straightforward methodology when training new staff members. Whilst we did not investigate the results–LIMS (Laboratory Information Management System) interface capability during this study, it can be envisaged that this could easily be accommodated, given that the results can be exported via a csv file or an Excel file. The identical PerkinElmer full IVD solution (EONISTM system) comes with a preinstalled analysis software that is compatible with the LIMS systems utilised in the UK, enabling full sample tracking from punching to result interpretation. We can also now suggest that assay QC can be assessed by using the Cq of the kit controls. Any run where control Cq lies outside of the 2 SD acceptability range could be considered void.
The hurdle of using DNA for NBS in the UK has already been partially overcome with the previous proof-of-concept study for SCID (severe combined immunodeficiency) NBS [
10] and the recently started UK SCID NBS pilot. The method used here for SMA NBS is already being used alongside SCID NBS in many countries and can be combined as a multiplex test [
9], adding very little to lab workload or cost. Hands-on time using this SMA screening assay alone was 2 h, which included punching, extracting DNA, assay setup, and analysis. Unlike SCID screening, which is a quantitative assay, SMA screening is qualitative and therefore not subject to cut-off values for result interpretation, which permitted a rapid analysis. If combined with SCID screening as a multiplex assay approach, the only extra hands-on time required would be for analysis, and this may be considered in future should SMA NBS screening be implemented in the UK. Discussion around screening for
SMN2 testing in the UK is not yet underway. Unlike for SCID screening where prematurity can be an issue affecting quantitation, this SMA assay is purely qualitative for the presence/absence of
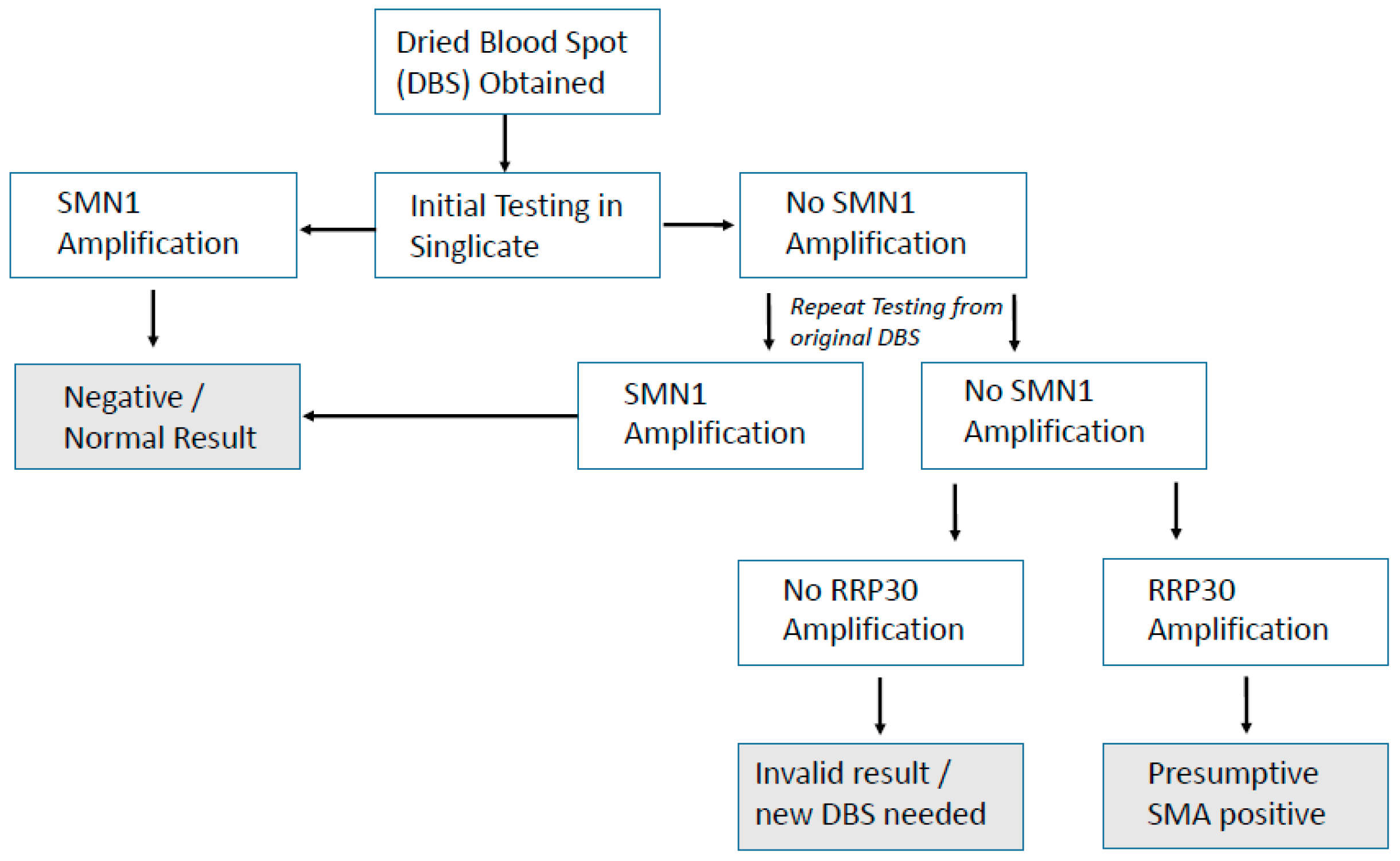
SMN1 exon 7. An algorithm already exists in the UK for SCID NBS, and a suggested simple algorithm for SMA screening is presented in
Figure 3.
In summary, our proof-of-concept study demonstrates that a simple commercially available rqPCR kit can be used in the routine clinical laboratory setting within the UK infrastructure for the detection of the exon 7 deletion in the SMN1 gene. It is simple to use and robust and provides a direct NBS test for downstream diagnostic assessment.
Author Contributions
Conceptualisation, S.P.A., M.S., S.S. and F.M.; methodology, S.P.A., E.G., N.K., S.K. and A.I.; validation, S.P.A., E.G., N.K., S.K. and A.I.; formal analysis, S.P.A., E.G., N.K. and S.K.; data curation, S.P.A., E.G., N.K. and S.K.; writing—original draft preparation, S.P.A.; writing—review and editing, S.P.A., M.S. and F.M.; visualization, S.P.A. and F.M.; supervision, S.P.A. and F.M.; project administration, S.P.A. and S.S.; funding acquisition, F.M. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
All research at Great Ormond Street Hospital NHS Foundation Trust and the UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre.
Institutional Review Board Statement
The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the London–West London and GTAC Research Ethics Committee (19/LO/1801).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available on request.
Acknowledgments
We acknowledge the support from both Biogen and PerkinElmer with supply of control material and reagents, respectively.
Conflicts of Interest
F.M. has received honoraria for presentation at meetings and scientific advisory boards of Biogen, Novartis, and Roche. He also acknowledges a grant from Biogen for the SMA REACH UK registry for patients affected by SMA. M.S. has received honoraria for presentation at meetings and scientific advisory boards of Biogen, Novartis, and Roche. Other authors declare no conflicts of interest.
References
- Verhaart, I.E.; Robertson, A.; Leary, R.; McMacken, G.; Konig, K.; Kirschner, J.; Jones, C.C.; Cook, S.F.; Lochmuller, H. A multi-source approach to determine SMA incidence and research ready population. J. Neurol. 2017, 264, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Verhaart, I.E.; Robertson, A.; Wilson, I.J.; Aartsma-Rus, A.; Cameron, S.; Jones, C.C.; Cook, S.F.; Lochmuller, H. Prevalence, incidence and carrier frequency of 5q–linked spinal muscular atrophy—A literature review. Orphanet J. Rare Dis. 2017, 12, 124–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Office of National Statistics. Births in England and Wales. 2019. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/livebirths (accessed on 6 September 2021).
- Kraszewski, J.N.; Kay, D.M.; Stevens, C.F.; Koval, C.; Haser, B.; Ortiz, V.; Albertorio, A.; Cohen, L.L.; Jain, R.; Andrew, S.P.; et al. Pilot study of population-based newborn screening for spinal muscular atrophy in New York state. Genet. Med. 2018, 20, 608–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandsema, J.; Gross, B.N.; Matesanz, S.E. Diagnostic testing for patients with spinal muscular atrophy. Clin. Lab. Med. 2020, 40, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.-H.; Chiang, S.-C.; Weng, W.-C.; Lee, N.-C.; Lin, C.-J.; Hsieh, W.-S.; Lee, W.-T.; Jong, Y.-J.; Ko, T.-M.; Hwu, W.-L. Presymptomatic diagnosis of spinal muscular atrophy through newborn screening. J. Pediatr. 2017, 190, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Boemer, F.; Caberg, J.-H.; Dideberg, V.; Dardenne, D.; Bours, V.; Hiligsmann, M.; Dangouloff, T.; Servais, L. Newborn screening for SMA in Southern Belgium. Neuromuscul. Dis. 2019, 29, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Vill, K.; Kolbel, H.; Schwartz, O.; Blaschek, A.; Olgemoller, B.; Harms, E.; Burggraf, S.; Roschinger, W.; Durner, J.; Glaser, D.; et al. One Year of Newborn Screening for SMA–Results of a German Pilot Project. J. Neuromuscul. Dis. 2019, 6, 503–515. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-Mateo, C.; Timonen, A.; Vaahtera, K.; Jaakkola, M.; Houggard, D.M.; Bybjerg-Grauholm, J.; Baekvad-Hansen, M.; Adamsen, D.; Filippov, G.; Daillaire, S.; et al. Development of a Multiplex Real-Time PCR Assay for the Newborn Screening of SCID, SMA, and XLA. Int. J. Neonatal Screen. 2019, 5, 39. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.P.; Rashid, S.; Premachandra, T.; Harvey, K.; Ifederu, A.; Wilson, M.; Gaspar, B. Screening of Neonatal UK Dried Blood Spots Using a Duplex TREC Screening Assay. J. Clin. Immunol. 2014, 34, 323–330. [Google Scholar] [CrossRef]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).